Review Article - Modern Phytomorphology ( 2023) Volume 17, Issue 1
Mitochondrial fatty acid synthesis in plants: enzymes and biological roles
L. Sarvananda1,2*, B.I.L.M. Mendis1, H.D.T. Madhuranga3 and Amal D Premarathna42Molecular Nutritional and Biochemistry Laboratory, University of Peradeniya, Peradeniya, Sri Lanka
3Department of Biomedical Science, Faculty of Health Science, KIU, Battaramulla, Sri Lanka
4School of Natural Sciences and Health, Tallinn University, Narva mnt 29, 10120 Tallinn, Estonia
L. Sarvananda, Department of Basic Veterinary Sciences, Faculty of Veterinary Medicine and Animal Science, University of Peradeniya, Peradeniya, Sri Lanka, Email: sarvacool18@gmail.com
Received: 15-Feb-2023, Manuscript No. mp-22- 89447; Accepted: 28-Feb-2023, Pre QC No. mp-22- 89447(PQ); Editor assigned: 18-Feb-2023, Pre QC No. mp-22- 89447(PQ); Reviewed: 21-Feb-2023, QC No. mp-22- 89447(Q); Revised: 25-Feb-2023, Manuscript No. mp-22- 89447(R); Published: 02-May-2023, DOI: 10.5281/zenodo.7939690
Abstract
Mitochondria are intracellular organelles located in the cytoplasm, that are essential for life and health. It is also known as the powerhouse of the cells and use a process called oxidative phosphorylation to generate energy. In side mitochondria, the lipid monomer fatty acids act as energy sources, membrane constituents, molecules for post translational modifications of proteins and cellular signaling factors. Plant cells possess couple of distinct type 2 Fatty Acid Synthase (FAS) systems. The two systems take place in plastids (ptFAS) and mitochondria (mtFAS). Lots of mtFAS components have been identified but, the physiological functions of mtFAS are still poorly understood. In this review, we have characterized five Arabidopsis mtFAS components, namely Mitochondrial β-Ketoacyl-ACP Synthase (mtKAS), Mitochondrial 3-Hydroxyacyl-ACP Dehydratase (mtHD), Mitochondrial Enoyl-ACP reductase (mtER), Mitochondrial Phosphopantetheinyl Transferase (mtPPT), and Mitochondrial Malonyl-CoA Synthetase (mtMCS). MtKAS, mtHD, and mtER catalyze 3 reactions of the 4 reactions of the acyl chain elongation cycle; mtPPT is responsible for activating Mitochondrial Acyl Carrier Protein (mtACP) by the addition of a phosphopantetheine cofactor; and mtMCS generates malonyl-CoA, the donor of 2-carbon elongation unit for the mtFAS system. Mitochondrial octanoyl-ACP is the precursor for the synthesis of lipoic acid, which is the coenzyme of the H-subunit of Glycine Decarboxylase Complex (GDC), a key enzyme of photorespiration. Accordingly, mutations in the mtFAS system can lead to a typical deficiency in photorespiration due to the depletion of the lipoylation of the H-subunit of GDC. These mutants are altered as an inferior effect of the deficiency in photorespiration. In addition, mutant analyses demonstrate that mitochondrial 3-hydroxymyristyl-ACP contributes to the biosynthesis of lipid alike molecules, suggesting a novel synthetic destination of the mtFAS intermediates. RNA-seq transcriptomic analyses of the mutants establish a regulatory network that is associated with the mtFAS functions is crucial for plant metabolic homeostasis.
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Haberler
Keywords
Mitochondrial fatty acid synthase, Mitochondrial acyl carrier proteins, Arabidopsis, fatty acid synthesis, Lipoic acid, Photorespiration, Malonyl Co A
Introduction
Mitochondria are intracellular organelles, that are essential for life and health, it is also known as the powerhouse of the cells and uses a process called oxidative phosphorylation to generate energy. The metabolism of living organisms is connected with a large variety of fatty acids that are originated from either exogenous sources or arising from endogenous synthesis (Kastaniotis A.J et al., 2017). Considering the mitochondria, fatty acids serve as, energy sources, membrane constituents, molecules for post translational modifications of proteins and signaling factors. The largest amount of fatty acid metabolism occurred through fatty acyl thioesters, which are more reactive than, sterol esters, phospholipids or triacylglycerols (Kastaniotis A.J et al., 2017). Fatty acids (FAS) are building blocks of acyl lipids, which are in the form of phospholipids or galactolipids. In the membranes or as TriGlycerol (TAG) oils, FAS also represents a major form of carbon storage in plant cells (Do T.H. et al, 2018; Li-Beisson Y. et al, 2017). Plant cells contained two distinct type 2 Fatty Acid Synthase (FAS) systems. These two systems occur and take place in plastids (ptFAS) and mitochondria (mtFAS) (Hiltunen J.K. et al, 2010). Type 2 FAS systems are contained with monofunctional enzymes that lead to production of fatty acids (Hiltunen J.K. et al, 2010). In contrast, the type 1 FAS system comprises multifunctional enzymes containing all the catalytic centers, which are essential to produce fatty acids (Guan X. et al., 2015). Type 1 FAS systems enzymes occur in the cytosol of fungi and animals. The ptFAS system produce the majority of fatty acids in a plant cell, as well as mtFAS and directly contribute to the biosynthesis of lipoic acid (Ewald R. et al, 2007). Acyl Carrier Protein (ACP) is central to all type 2 FAS systems, which connect acyl intermediates between the active sites of each enzyme component. These metabolic intermediates are covalently bound to the thioester bond halide with the posphopantetheine cofactor of holo ACP. As a result of that, ACP is initially expressed as an inactive apoproteins and it is activated by the Phosphopantetheinylation of a specific serine residue. This reaction is driven by the Phosphopantetheinyl Transferase (PPT) (White S.W. et al, 2005; Hiltunen J.K. et al, 2010). Therefore, each of the FAS system in plant cells has a dependency against a phosphopantetheinylation reaction, however, it has not as yet been well characterized (White S.W. et al, 2005).
In humans, lipoate is an essential cofactor for five redox reactions: four 2 oxoacid dehydrogenases and the Glycine Cleavage System (GCS). In addition to the two enzymes that are important for energy metabolism, branched chain ketoacid dehydrogenase, 2 oxoadipate dehydrogenase, and the GCS, three enzymes are also important for amino acid metabolism (Mayr J.A. et al., 2014). There is a similar architecture to all of these enzymes, and they all contain multiple subunits. There are three steps involved in lipoate synthesis in mitochondria, including mitochondrial fatty acid synthesis up to an octanoyl-acyl carrier protein, Lipoyl(octanoyl) Transferase 2 (putative) (LIPT2) transfer of octanoic acid to the glycine cleavage H protein, Lipoyltransferase 1 (LIPT1) synthesis of lipoate, Lipoyltransferase 1 (LIPT1), which lipoylates E2 subunits of 2-oxoacid dehydrogenases (Mayr J.A. et al., 2014).
There are three families of PPT enzymes have been characterized to date, primarily from microbial sources. Family 1 PPT is typically from E-coli; Family 2 PPT is from Bacillus subtilis, and Family 2 PPT are more promiscuous; Family 3 PPTs reside only in fungi, sharing sequence homology with Family 1 PPT, but they are specific for Type 1 FAS. Eukaryotic PPTs have been well characterized in the yeast Saccharomyces cerevisiae and humans (Guan X. et al., 2015). As well as Acyl carboxylates are activated by the process acylation via a thioester bond to coenzyme A(CoA). The resulting acyl-CoA are metabolic intermediates with high energy, that are used to production of fatty acids and amino acids (Guan X.et al., 2016). The importance of these intermediates in plant metabolism may be reflected with large number of Acyl-CoA synthetases that are found in the plant genomes. Malonyl-CoA is one such important metabolic intermediate and it has many metabolic processes; however, it functions on the two carbon donor substrate in the extension reactions of polyketide and fatty acid biosynthesis (Li-Beisson Y. et al., 2013). In this review, we have mainly focused on Mitochondrial Acyl Carrier Protein (mtACP) and its activation, mitochondrial acyl chain elongation cycle; and mitochondrial malonyl ACP, the donor of 2-carbon units; and also lipoic acid and its functions in photorespiration. In addition, a separate discussion on molecules which are like a lipid A; and as well as the plant metabolomics methodology is included which supports the investigation of the physiological significance of Arabidopsis mtFAS system.
Mitochondrial acyl carrier protein (mtACP) phylogenetic context, their expression, structure and its activation
At least three enzymes genetic fatty acids in plants, and these are used to produced cellular lipids such as glycerolipids in membrane and triacylglycerol in deeds. These are produced by plastid localized Type 2 FAS (ptFAS), it gives rise to 16 carbon-18 carbon atoms containing acyl chains. Further they enter in to the endoplasmic reticulum localized Fatty Acid Elongate system (FAE) to develop very long FA. Mitochondria generate Lipoic acid through type 2 FAS. It essential for many enzymes as an essential cofactor (Li-Beisson Y. et al., 2013). Acyl Carrier Protein (ACP) is a principal partner in the cytosolic and mitochondrial fatty acid synthesis pathways. ACP is a highly consumed cofactor that is essential for Type 2 Fatty Acid Synthases (FASs). Each FAS system utilizes ACP as a cofactor, that shuttles acyl intermediates between active sites of the catalytic components of each FAS system. the ACP dependent functions cannot achieved without converting the inactive apo foam into the active holo form via the post transitional phosphopantetheinylation of consumed residue. The terminal thiol of the ACP carried phosphopantetheinearm covalently binds to the acyl via high energy thioester bond. (Fig.1) shows the wider phylogenetic context of the Arabidopsis ACPs in relation to ACPa from 39 flowering plants. mtACP genes were characterized using RNA extracted from several tissues of wild type plants using the RT-qPCR. The results showed the relative transcript abundance of the three genes mtACP1, mtACP2, and mtACP3 across the seedlings, leaves, roots, stem, flowers, siliques (Fig.2). These expression patterns of mt ACPs are consistent with public microarray and RNAseq data and are similar to the expression of other mtFAS gene components. The study analysis was done using confocal micrographs of the roots of the used transgenic plants as compared to control plants. The GFP fluorescence showed all three mtACP fusions colocalized with mitochondrial marker, MitoTraker. Those analyses, therefor confirm the mitochondrial localization of the mtACP1, mtACP2, and mtACP3 proteins( Fu X. et al., 2020) (Fig.3).
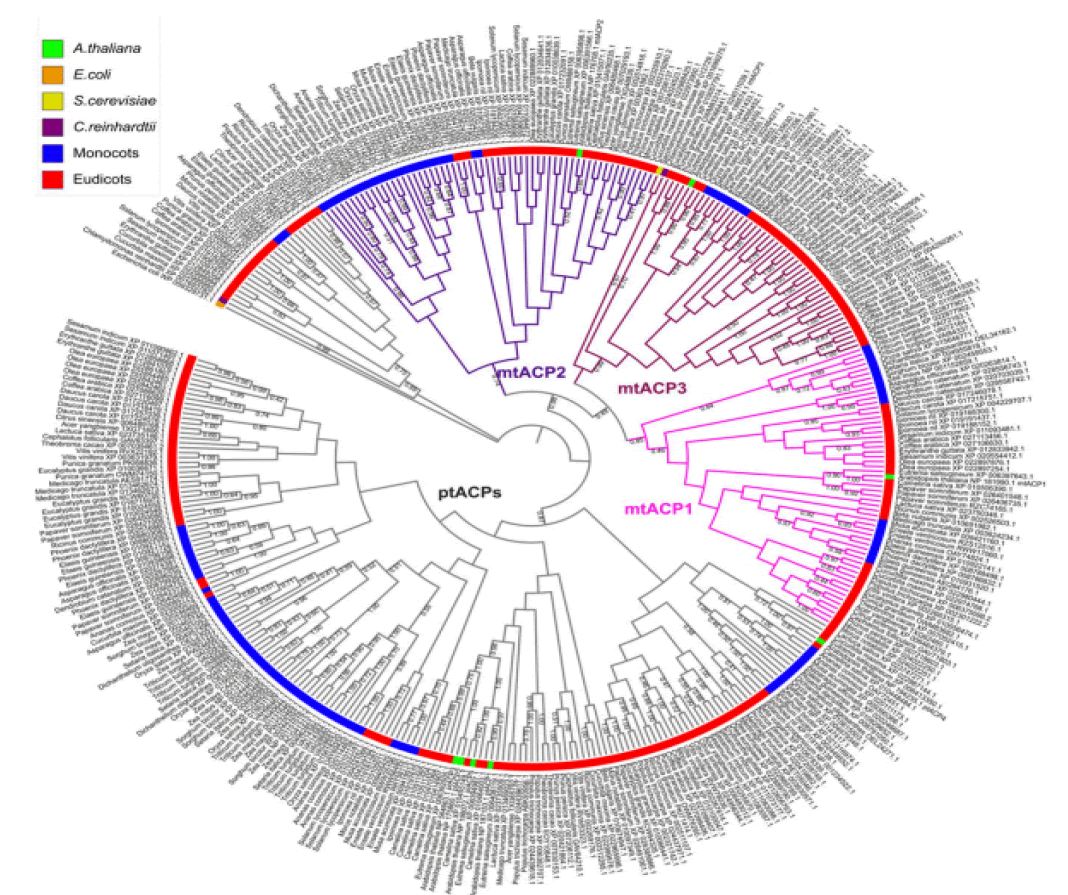
Figure 1: Phylogenetic analysis of ACPs in relation to ACPa from 39 flowering plants
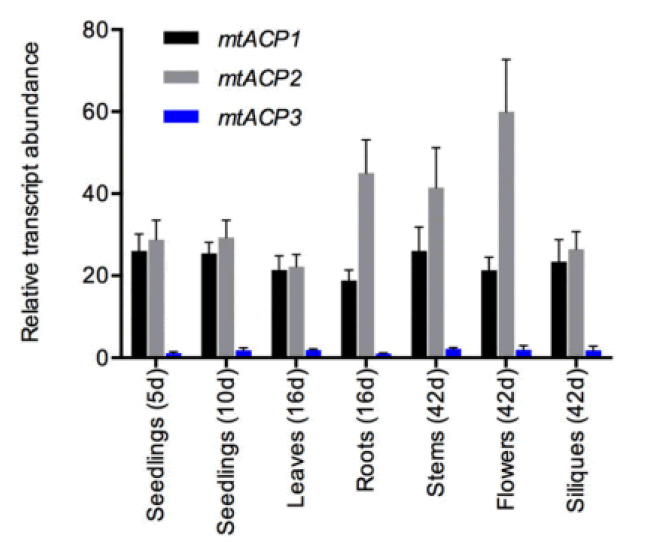
Figure 2: Relative transcript abundance of the three genes mtACP-1, mtACP-2, and mtACP-3 of the seedlings, leaves, roots, stems, flowers and
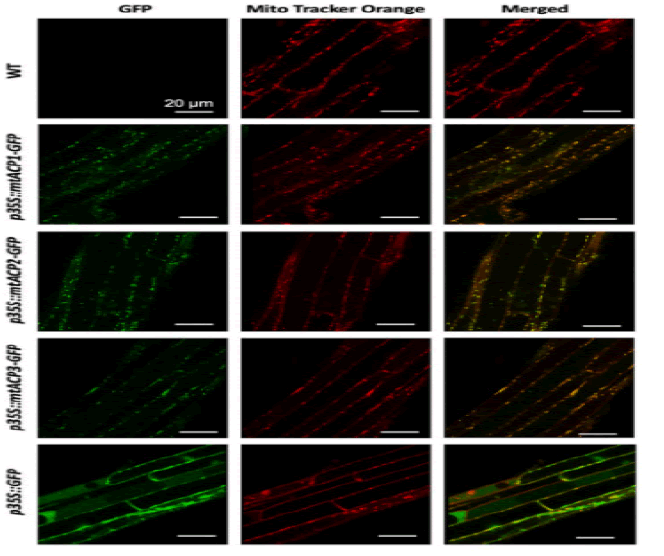
Figure 3: Sub cellular localization of Arabidopsis mtACP-1. mtACP-2, and mtACP-3
ACPs evolutionarily conserved small α helical proteins. That present acyl chain intermediates to catalytic sites of enzymes in the fatty acid synthesis pathway. In either case, ACP carries a 4’–phosphopantetheine (4’-PP) moiety covalently attached to a conserved serine with thioester bond. The number of amino acids residues in the shown ACPs are as follows. Study indicated “H. sapiens, 156; D. melanogaster, 152; X. laevis, 149; M. musculus, 156; S. cerevisiae, 125; A. thaliana, 126; E. coli, 78; P. falciparum, 121. The N-terminal amino acid sequences of ACPs are not shown and the given numbering follows the mature form of human ACP(NDUFAB1) (Masud A.J. et al., 2019).The α-helical secondary structure elements and residue numbers above correspond to the human mitochondrial ACP (PDB ID 5XTD, chain G/X) from the EM structure of respiratory complex I and was generated using ESPript 3.0. The key conserved residues are highlighted in red (Fig.4A). The 4α-PP attachment residue serine is indicated by a black arrow. TTT represent strict α–turns. Notably, the two negatively charged amino acids aspartic acid (D) and glutamic acid (E) in helix II that have been implied in the interaction with cognate enzymes and other partner proteins are well conserved across the different species. (Fig.4B) Schematic chemical formula of holo-ACP. Here, an acylated 4α-PP moiety is covalently bound to the hydroxyl group of an invariant serine residue of ACP (Ser112 in human) (Masud A.J. et al., 2019).
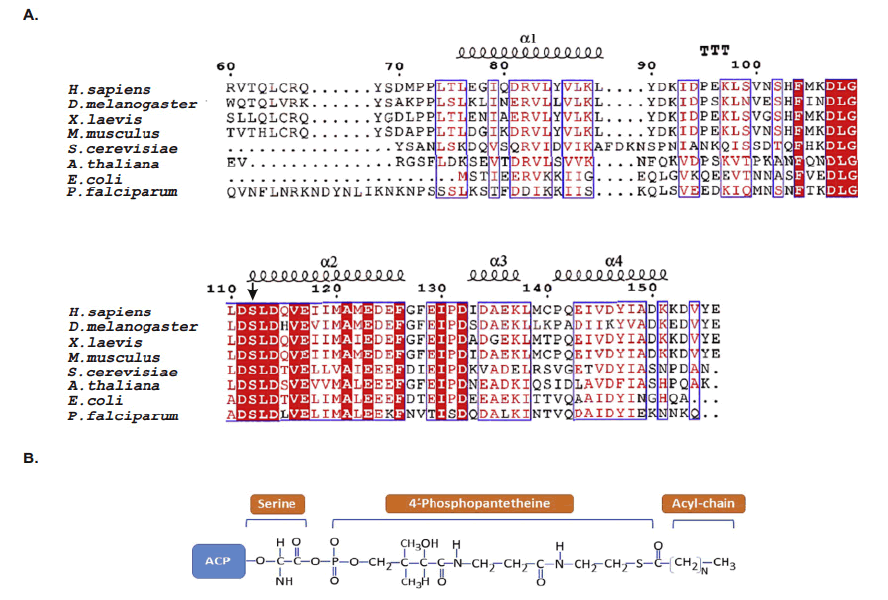
Figure 4: Multiple sequence alignment of ACPs
Mature ACP contains four α helices 1, 2, 3 and 4 connected by three interspersed loops and more structural features of mitochondrial ACP can be introduced as follows (Masud A.J. et al., 2019) (Fig.5). Primary reaction of Non-Ribosomal Peptide Synthetase(NRPS), PKS, and FAS by PPTases. PPTases transfer the 4’–phosphopantetheinyl moiety of coenzyme A to the side chain hydroxyl of a serine residue invariant in Peptidyl Carrier Protein domain (PCP) and ACPs. Thus, the carrier proteins are converted from the inactive apo form into the active holo form. It can introduced as followed schematic representation (Mofid M.R. et al., 2002). During fatty acid synthesis, the fatty acyl chain needs to be attached to the holo-ACP at the prosthetic 4’-phosphopantetheinyl moiety. However, newly synthesized ones are lack of it. The phosphopantetheinyl moiety is post translationally transferred from CoA to a ser residue in the ACP by a Phospantetheinyl transferase (PPTase) (EC 2.7.8.7) (Fig.6). There are two major types of PPTases. The holo-ACP Synthase (ACPS)-type PPTases are typically significant to the type 2 FAS or Polyketide Synthase (PKS) in Bacteria, e.g. Escherichia coli AcpS. The Surfactin Production Element (SFP)-type enzymes are mostly specific to type 1 FAS,PKS, or NRPS, such as the Bacillus subtilis SFP, Yeast Lys5, and human PPTase ( Joshi A.K. et al., 2003). The FAS α-subunit (FAS2) in fungi also contains an ACPS-type PPTase domain and is sometimes referred to as a third type of PPTase ( Mootz H.D. et al., 2002) (Fig.7).
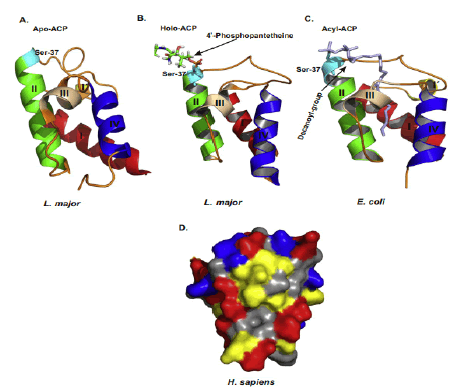
Figure 5: Structures of ACP and surface charge distribution
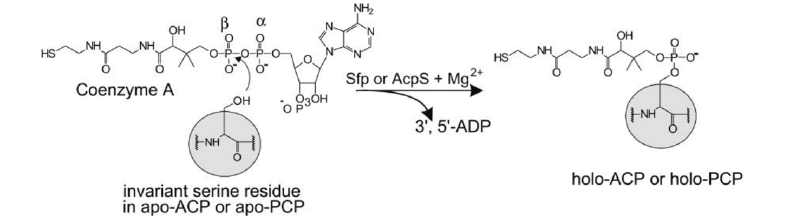
Figure 6: Priming reaction of NRPS, PKS, and FAS by PPTases.
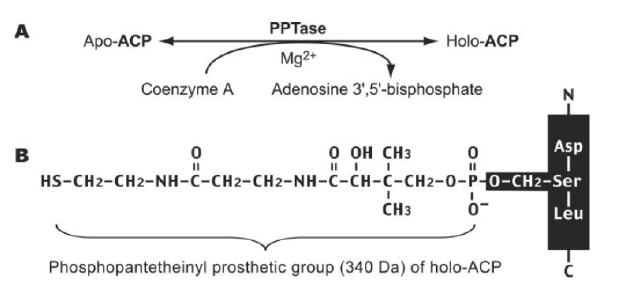
Figure 7: (A) PPTases synthesize holo-ACP by transferring the phosphopantetheinyl moiety from CoA to apo-ACP. (B) the Phosphopantetheinyl moiety is attached to the ser residue of ACP.
Mitochondrial acyl chain elongation cycle
The iterative mitochondrial fatty acid elongation cycle is catalyzed by 4 enzymes, namely Mitochondrial α-Ketoacyl-ACP Synthase (mtKAS), Mitochondrial α-Ketoacyl-ACP Reductase (mtKR), Mitochondrial 3-Hydroxyacyl-ACP Dehydratase (mtHD), and Mitochondrial Enoyl-ACP Reductase (mtER). MtKAS supports the condensation reaction of the fatty acyl chain elongation cycle. In plants, mtKAS has been previously characterized in Arabidopsis (Venkatesan R. et al., 2014). Mitochondrial Fatty Acid Synthesis (mtFAS) is essential for respiratory growth in yeast and mammalian embryonic survival. The human 3-Ketoacyl-Acyl Carrier Protein (ACP) Reductase (KAR) of mtFAS is a heterotetrameric a2b2-assembly composed of 17b-Hydroxysteroid Dehydrogenase Type-8 (HSD17B8, a-subunit) and Carbonyl Reductase Type-4 (CBR4, b-subunit). Here we provide a structural explanation for the stability of the heterotetramer from the crystal structure with NAD þ and NADP þ bound to the HSD17B8 and CBR4 subunits, respectively, and show that the catalytic activity of the NADPH and ACP-dependent CBR4 subunit is crucial for a functional HsKAR. Therefore, mtFAS is NADPHand ACP dependent, employing the 3R-hydroxyacyl-ACP intermediate. HSD17B8 assists in the formation of the competent HsKAR assembly. The intrinsic NAD þ- and CoA-dependent activity of the HSD17B8 subunit on the 3R-hydroxyacyl-CoA intermediates may indicate a role for this subunit in routing 3R-hydroxyacyl-CoA esters, potentially arising from the metabolism of unsaturated fatty acids, into the mitochondrial b-oxidation pathway (Venkatesan R. et al., 2014) (Fig.8).
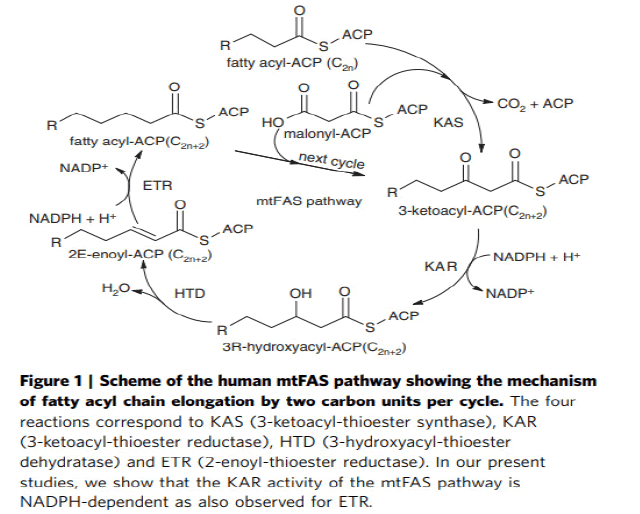
Figure 8: Mitochondrial acyl chain elongation cycle
Mitochondrial malonyl-ACP, the donor of 2-carbon elongation units
A study was demonstrated that the malonyl-CoA synthetase enzyme, previously identified as being encoded by the Arabidopsis AAE13 gene, generates the mitochondrial malonyl-CoA pool by ligating malonate to CoA (Guan X.et al., 2016). This result is consistent with a previous finding that malonyl-CoA synthetase activity is measurable in the soluble fraction of purified pea mitochondria. Furthermore, this finding involving the Arabidopsis AAE13 protein can be extrapolated generally among other higher plant species because the N-terminal mitochondrion-targeting leader presequence that occurs in the AAE13 protein also occurs in orthologs of higher plant species whose genomes have been sequenced. Thus, in silico predictions indicate that plant mitochondria in general can generate malonyl-CoA in organello from malonate (Guan X.et al., 2016). This reflects the situation that occurs in human mitochondria, where malonyl-CoA is similarly assembled from malonate via the reaction catalyzed by the ACSF3 protein, a malonyl-CoA synthetase. In contrast, however, yeast mitochondria generate malonyl-CoA by the carboxylation of Acetyl-CoA, catalyzed by a homomeric ACCase isoform (Hfa1p). Similarly, mitochondrially located ACCase has been reported in Poaceae and rice via proteomic analyses. However, additional genomic characterizations have not validated these findings of a mitochondrial ACCase (Guan X.et al., 2016; Bowman C.E. et al., 2019) (Fig. 9).
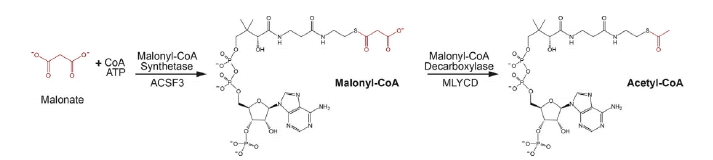
Figure 9: Coordinated metabolism of malonyl CoA by ACSF3 and MYLCD
Lipoic acid and its functions in photorespiration
Lipoic acid is almost universally required for aerobic metabolism. However, the mechanism for its synthesis and incorporation into proteins has remained elusive. Therefore, having an idea about lipoic acid biosynthesis is very important. When you consider about lipoic acid biosynthesis pathway, not only plant but some species have several paths for synthesis of lipoic acid. in the cell very little lipoate exist as the free acid, almost all is tethered to the L-amino acid lipoyl attraction group. Pioneering studies from Crohn's laboratory indicate that E coli maintain at least two pathways for attaching the lipoyl group to these target lysine residues. Lipoic acid pathway can be introduced as the follows (Booker S.J, 2004). In addition, (Fig. 10) showed that metabolism or utilization of lipoic acid. In plants, lipoic acid biosynthesis is associated with photorespiration, which recruits Glycine Decarboxylase Complex (GLD) containing a lipoylated H-protein subunit. Photorespiration occurs in all oxygen-producing photosynthetic organisms, including plants, algae, and cyanobacteria. Photorespiration is initiated by the oxygenase activity of Ribulose-1,5-BisphosphateCarboxylase/Oxygenase (RUBISCO), the same enzyme that is also responsible for CO2 fixation in almost all photosynthetic organisms. Phosphoglycolate formed by oxygen fixation is recycled to the Calvin cycle intermediate phosphoglycerate in the photorespiratory pathway. This reaction cascade consumes energy and reducing equivalents and part of the afore fixed carbon is again released as CO2 (Peterhansel C. et al., 2010). Because of this, photorespiration was often viewed as a wasteful process. Here, we review the current knowledge on the components of the photo respiratory pathway that has been mainly achieved through genetic and biochemical studies in Arabidopsis. Based on this knowledge, the energy costs of photorespiration are calculated, but the numerous positive aspects that challenge the traditional view of photorespiration as a wasteful pathway are also discussed. An outline of possible alternative pathways beside the major pathway is provided (Peterhansel C. et al., 2010) (Fig. 11).
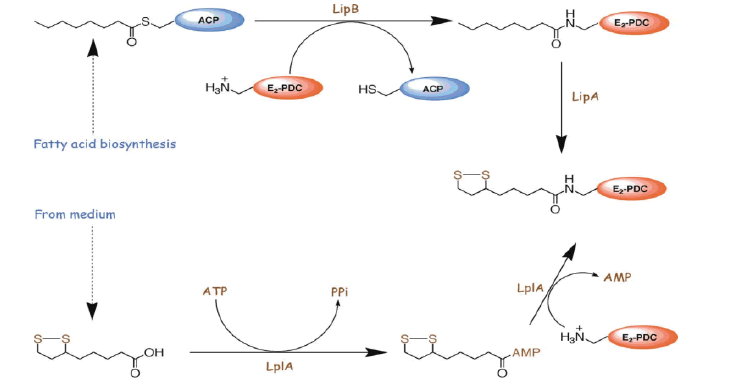
Figure 10: Lipoic acid biosynthesis pathway
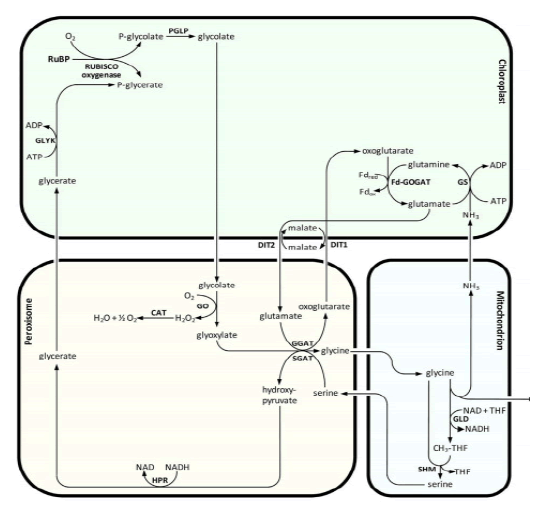
Figure 11: The major photorespiratory pathway in Arabidopsis
Lipid molecule
A lipid is the hydrophobic anchor of the outer monolayer of outer membrane of most gram-negative bacteria. Arabidopsis mitochondria are characterized to contain orthologs (e.g., AtLpxA, AtLpxC, AtLpxD1, AtLpxD2, AtLpxB, AtLpxK, and AtKdtA) of E.coli enzymes of lipid A biosynthesis. Lipid X, an intermediate of lipid biosynthesis that contains two 3-hydroxymyristyl chains on the sugar molecule, has been detected in the wild-type Arabidopsis plants. The contents of lipid X are diminished when genes encoding either of the two acyl transferases (i.e., AtLpxA and AtLpxD1) are deleted, or when the expression of AtLpxC is suppressed. Mutant analysis also supports the continuation of this synthetic pathway beyond lipid X, namely AtLpxB catalyzed formation of tetra acyl disaccharide phosphate, AtLpxK-catalyzed addition of a second phosphate group, and AtKdtA-catalyzed addition of two Kdo-moieties. To date, the final structure and physiological functions of lipid A like molecules have yet to be discovered in plants ( Li C., et al., 2011; Guan X. et al., 2017; Guan X. et al., 2020).
Plant metabolomics methodology
The total number of metabolites in the plant kingdom is estimated to range from 200,000 to 1,000,000 (Dixon R.A, 2003). A single accession of Arabidopsis thaliana is expected to produce about 5,000 metabolites. 2,632 of which are contained in AraCyc version 6.0 (Swarbreck D. et al., 2007). Because metabolites are the final products of genes and proteins, plant metabolome (a complete set of metabolites) represents the ultimate phenotype in response to genetic or environmental alterations (Saito K. et al., 2010). Metabolites bear a large dynamic range (~10^6) in the relative concentration and the various biochemical properties and, thus, comprehensive coverage can be only achieved by using multi parallel complementary analytical platforms. Indeed, a combination of eight different mass spectrometry-based platforms successfully leads to the quantification of about 2,000 metabolites in Arabidopsis (Quanbeck S.M. et al., 2012). To understand the cellular alterations as a consequence of the deficiency in mtFAS, we analyze the mtFAS mutants using 3 analytical platforms in this dissertation, namely HPLC, GC-MS, and LC-MS. HPLC method is sensitive in amino acids profiling when combined with OPA-based fluorescence derivatization for the amino group (Pereira V. et al., 2008). GC-MS method is able to detect primary metabolites, such as hydrocarbons, organic acids, and sugars by employing chemical derivatization for the hydrophilic groups (Lisec J. et al., 2006). LC-MS is efficient in the detection of small polar metabolites, but this method can be also used for the profiling of lipids when utilizing different columns and mobile phases (Okazaki Y.et al., 2009).
Conclusion
The Arabidopsis mitochondrial fatty acid synthase (mtFAS) include mitochondrial α- Ketoacyl-ACP Synthase (mtKAS), Mitochondrial 3-Hydroxyacyl-ACP Dehydratase (mtHD), Mitochondrial Enoyl-ACP Reductase (mtER), Mitochondrial Phosphopantetheinyl Transferase (mtPPT), and Mitochondrial Malonyl-CoA Synthetase (mtMCS). MtKAS, mtHD, and mtER catalyze 3 reactions of the 4 reactions of the fatty acyl chain elongation cycle; mtPPT supports the activation of apo-mtACP isoforms; and mtMCS generates malonyl-CoA, the precursor for the acyl chain elongation process. These monofunctional enzymes support fatty acid biosynthesis, which mimics the canonical Type II FAS in bacteria and plant plastids. The biochemical and physiological characterization of these enzymes demonstrate the occurrence of the mtFAS system in Arabidopsis and its functional significance to plant growth and development.
References
Booker S.J (2004). Unraveling the pathway of lipoic acid biosynthesis. Chem biol. 11: 10-12. [Google Scholar] [Cross Ref]
Bowman C.E., Wolfgang M.J (2019). Role of the malonyl-CoA synthetase ACSF3 in mitochondrial metabolism. Adv biol regul. 71: 34-40. [Google Scholar] [Cross Ref]
Dixon R.A (2003). Phytochemistry meets genome analysis, and beyond. Phytochemistry.; 62: 815-816. [Google Scholar] [Cross Ref]
Do T.H., Martinoia E., Lee Y (2018). Â Functions of ABC transporters in plant growth and development. Curr opin plant biol. 41: 32-38. [Google Scholar] [Cross Ref]
Ewald R., Kolukisaoglu U., Bauwe U., Mikkat S., Bauwe H (2007). Mitochondrial protein lipoylation does not exclusively depend on the mtKAS pathway of de novo fatty acid synthesis in Arabidopsis. Plant physiol. 145: 41-48. [Google Scholar] [Cross Ref]
Fu X., Guan X., Garlock R., Nikolau B.J (2020). Mitochondrial fatty acid synthase utilizes multiple acyl carrier protein isoforms. Plant Physiology. 183: 547-557. [Google Scholar] [Cross Ref]
Guan X., Chen H., Abramson A., Man H., Wu J., Yu O., Nikolau B.J (2015). A phosphopantetheinyl transferase that is essential for mitochondrial fatty acid biosynthesis. Plant J. 84: 718-732. [Google Scholar] [Cross Ref]
Guan X., Nikolau B.J (2016). AAE 13 encodes a dualâ?localized malonylâ?CoA synthetase that is crucial for mitochondrial fatty acid biosynthesis. Plant J. 85: 581-593. [Google Scholar] [Cross Ref]
Guan X., Okazaki Y., Lithio A., Li L., Zhao X., Jin H., Nettleton D., Saito K., Nikolau B.J (2017). Discovery and characterization of the 3-hydroxyacyl-ACP dehydratase component of the plant mitochondrial fatty acid synthase system. Plant physiol. 173: 2010-2028. [Google Scholar] [Cross Ref]
Guan X., Okazaki Y., Zhang R., Saito K., Nikolau B.J (2020). Dual-localized enzymatic components constitute the fatty acid synthase systems in mitochondria and plastids. Plant physiol. 183: 517-529. [Google Scholar] [Cross Ref]
Hiltunen J.K., Chen Z., Haapalainen A.M., Wierenga R.K., Kastaniotis A.J (2010). Mitochondrial fatty acid synthesisâ??an adopted set of enzymes making a pathway of major importance for the cellular metabolism. Prog lipid res. 49: 27-45. [Google Scholar] [Cross Ref]
Joshi A.K., Zhang L., Rangan V.S., Smith S (2003). Cloning, expression, and characterization of a human 4â?²-phosphopantetheinyl transferase with broad substrate specificity. J Biol Chem. 278: 33142-33149. [Google Scholar] [Cross Ref]
Kastaniotis A.J., Autio K.J., Keratar J.M., Monteuuis G., Makela A.M., Nair R.R., Pietikainen L.P., Shvetsova A., Chen Z., Hiltunen J.K (2017). Mitochondrial fatty acid synthesis, fatty acids and mitochondrial physiology. Biochim Biophys Acta (BBA)-Mol Cell Biol Lipids. 1862: 39-48. [Google Scholar] [Cross Ref]
Li C., Guan Z., Liu D., Raetz C.R (2011). Pathway for lipid A biosynthesis in Arabidopsis thaliana resembling that of Escherichia coli. Proc Natl Acad Sci. 108: 11387-11392. [Google Scholar] [Cross Ref]
Li-Beisson Y., Neunzig J., Lee Y., Philippar K (2017). Plant membrane-protein mediated intracellular traffic of fatty acids and acyl lipids. Curr opin plant biol. 40: 138-146. [Google Scholar] [Cross Ref]
Li-Beisson Y., Shorrosh B., Beisson F., Andersson M.X., Arondel V., Bates P.D., Baud S., Bird D., DeBono A., Durrett T.P., Franke R.B (2013). Acyl-lipid metabolism. Arab book/Am Soc Plant Biol. 11: e0161 [Google Scholar] [Cross Ref]
Lisec J., Schauer N., Kopka J., Willmitzer L., Fernie A.R (2006). Gas chromatography mass spectrometryâ??based metabolite profiling in plants. Nat protoc. 1: 387-396. [Google Scholar] [Cross Ref]
Masud A.J., Kastaniotis A.J., Rahman M.T., Autio K.J., Hiltunen J.K (2019). Mitochondrial acyl carrier protein (ACP) at the interface of metabolic state sensing and mitochondrial function. Biochim Biophys Acta (BBA)-Mol. Cell Res. 1866: 118540. [Google Scholar] [Cross Ref]
Mayr J.A., Feichtinger R.G., Tort F., Ribes A., Sperl W (2014). Lipoic acid biosynthesis defects. J inherit metab dis. 37: 553-563. [Google Scholar] [Cross Ref]
Mofid M.R., Finking R., Marahiel M.A (2002). Recognition of hybrid peptidyl carrier proteins/acyl carrier proteins in nonribosomal peptide synthetase modules by the 4â?²-phophopantetheinyl transferases AcpS and Sfp. J Biol Chem. 277: 17023-17031. [Google Scholar] [Cross Ref]
Mootz H.D., Schorgendorfer K., Marahiel M.A (2002). Functional characterization of 4â?²-phosphopantetheinyl transferase genes of bacterial and fungal origin by complementation of Saccharomyces cerevisiae lys5. FEMS microbiol lett. 213: 51-57. [Google Scholar] [Cross Ref]
Okazaki Y., Shimojima M., Sawada Y., Toyooka K., Narisawa T., Mochida K., Tanaka H., Matsuda F., Hirai A., Hirai M.Y., Ohta H (2009). A chloroplastic UDP-glucose pyrophosphorylase from Arabidopsis is the committed enzyme for the first step of sulfolipid biosynthesis. Plant Cell. 21: 892-909. [Google Scholar] [Cross Ref]
Pereira V., Pontes M., Câmara J., Marques J.C (2008). Simultaneous analysis of free amino acids and biogenic amines in honey and wine samples using in loop orthophthalaldeyde derivatization procedure. J Chromatogr A. 1189: 435-443. [Google Scholar] [Cross Ref]
Peterhansel C., Horst I., Niessen M., Blume C., Kebeish R., Kürkcüoglu S., Kreuzaler F (2010). Photorespiration. Arab book. 2010. [Google Scholar] [Cross Ref]
Quanbeck S.M., Brachova L., Campbell A.A., Guan X., Perera A., He K., Rhee S.Y., Bais P., Dickerson J.A., Dixon P., Wohlgemuth G (2012). Metabolomics as a hypothesis-generating functional genomics tool for the annotation of Arabidopsis thaliana genes of “unknown function”. Front plant sci. 3: 15. [Google Scholar] [Cross Ref]
Saito K., Matsuda F (2010). Metabolomics for functional genomics, systems biology, and biotechnology. Annu rev plant biol. 61: 463-489. [Google Scholar] [Cross Ref]
Swarbreck D., Wilks C., Lamesch P., Berardini T.Z., Garcia-Hernandez M., Foerster H., Li D., Meyer T., Muller R., Ploetz L, Radenbaugh A (2007). The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic acids res. 36: 1009-1014. [Google Scholar] [Cross Ref]
Venkatesan R., Sah-Teli S.K., Awoniyi L.O., Jiang G., Prus P., Kastaniotis A.J., Hiltunen J.K., Wierenga R.K., Chen Z (2014). Insights into mitochondrial fatty acid synthesis from the structure of heterotetrameric 3-ketoacyl-ACP reductase/3R-hydroxyacyl-CoA dehydrogenase. Nat commun. 5: 4805. [Google Scholar] [Cross Ref]
White S.W., Zheng J., Zhang Y.M., Rock C.O (2005). The structural biology of type II fatty acid biosynthesis. Annu Rev Biochem. 74: 791-831. [Google Scholar] [Cross Ref]